HRD test
scarHRD and sequenza
Jun Kang
2021-11-08
Myriad myChoice CDx
- Homologous recombination deficiency (HRD) status
- Presence mutations in BRCA1 and BRCA2 genes
- Sensitivity to platinum-based chemotherapy
- Ovarian cancer
- Treatment with Zejula (niraparib)
Homologous recombination deficiency (HRD)
- Double-strand break
- Genomic scars
- Large-scale loss of heterogeneity [1]
- Telomere allelic imbalance [2]
- Large-scale state transition [3]
Loss of Heterozygosity (HRD-LOH)
- Number of 15 Mb exceeding LOH regions which do not cover the whole chromosome.
Large Scale Transitions (LST)
- Chromosomal break between adjacent regions of at least 10 Mb, with a distance between them not larger than 3Mb
Telomeric Allelic Imbalances
- Number AIs that extend to the telomeric end of a chromosome.
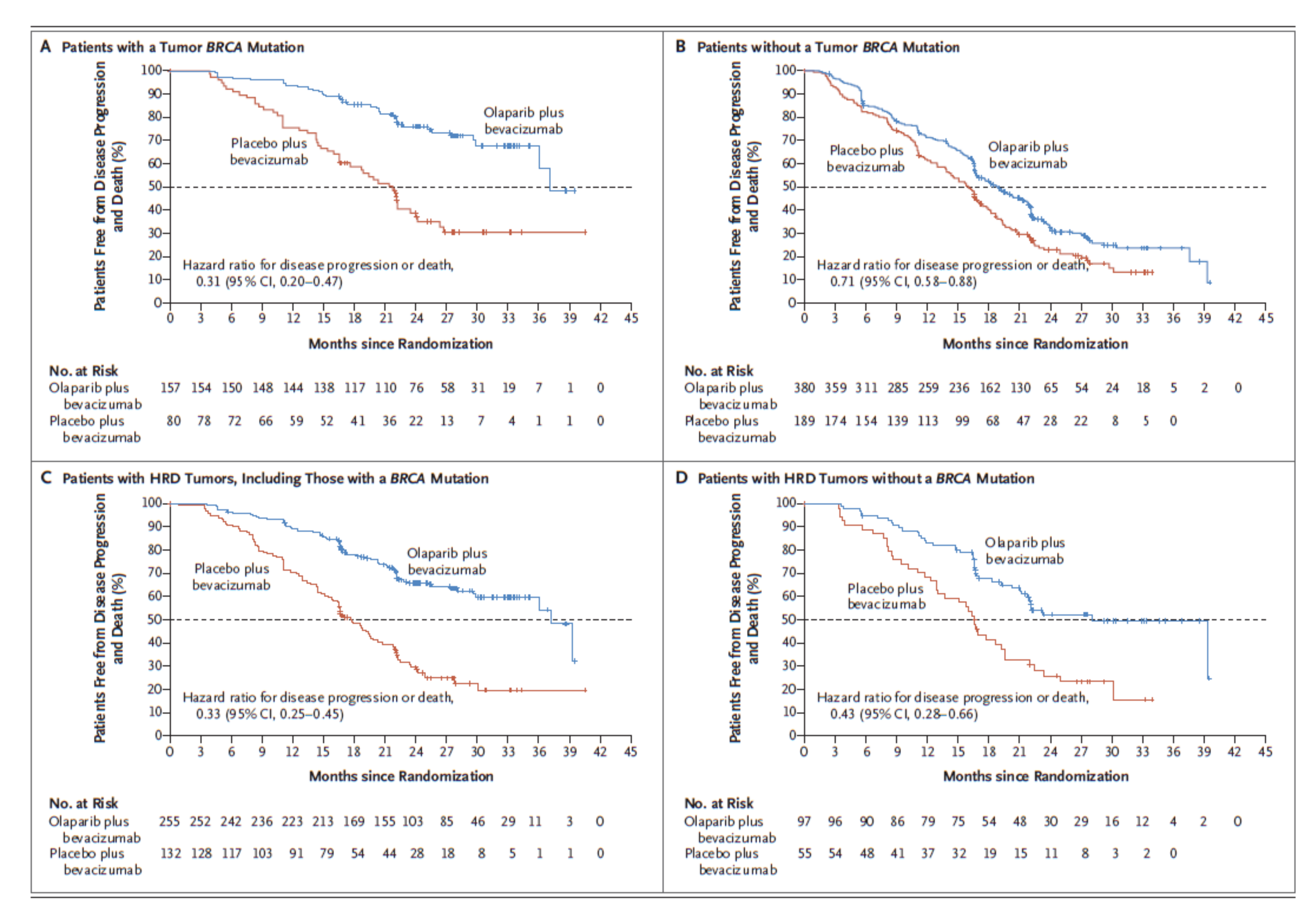
HRD test as a companion diagnostics [4]
Key concepts
- Allele-specific copy number
- Allele frequency of heterozygous single nucleotide polymorphism (heterozygous SNP)
HRD test programs
- Sequenza [5]
- scarHRD [6]
Allele-specific copy number analysis of tumors [7]

- B allele frequency (BAF): relative quantity of the one allele compared to the other
- log R ratio (LRR): Total probe intensity of a given SNP relative to a canonical set of normal controls
- Loss of heterozygosity
- Allelic imbalances
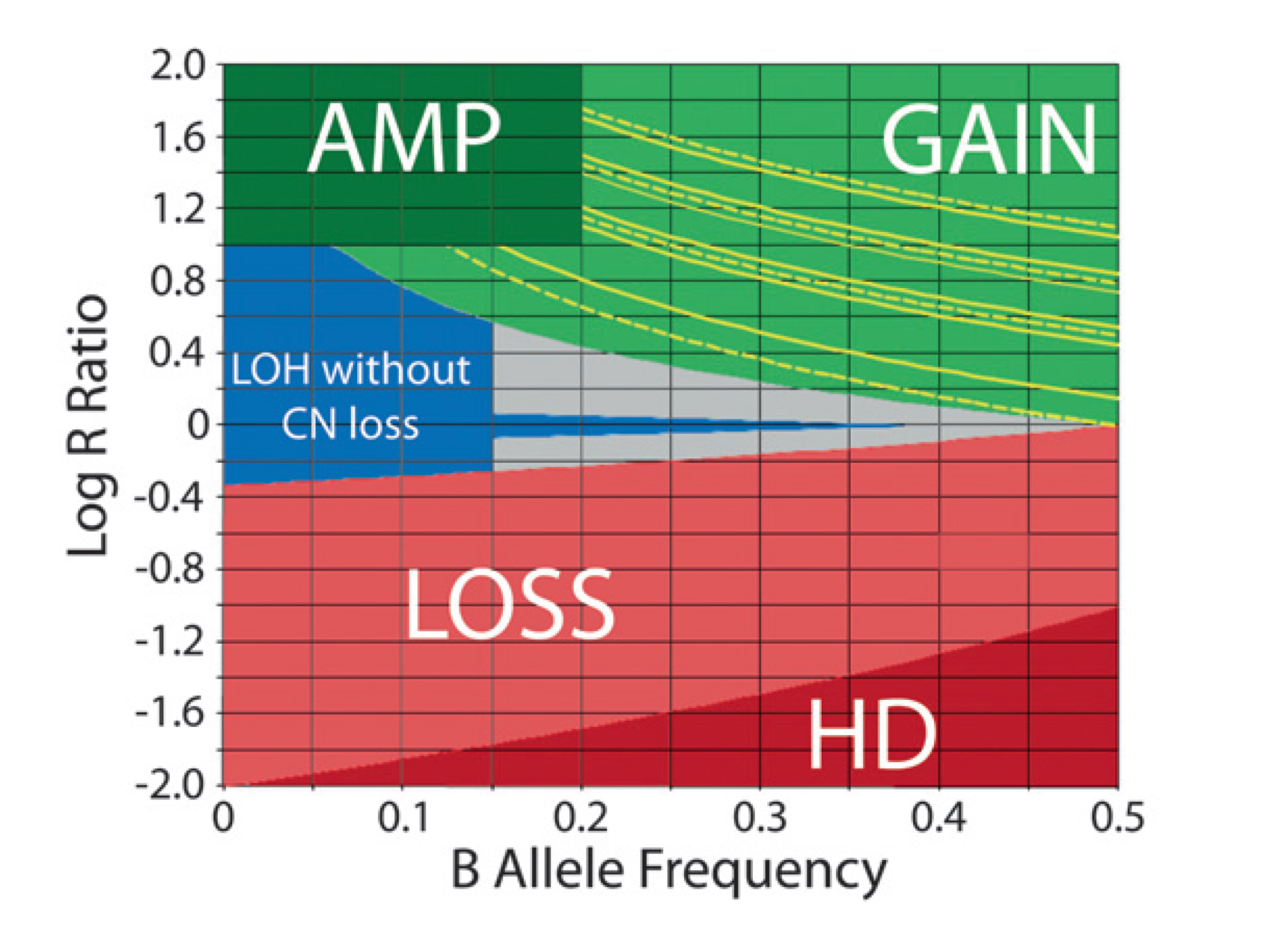
Allele frequency[8]
BAF = pV+1−ppC+2(1−p)
- Given copy number (C)
- Variant allele count (V)
- Sample purity (p)
Three steps of Bayesian data analysis
- Full probability model
- Conditioning on observed data (posterior distribution)
- Evaluating the fit of the model and the implications of the resulting posterior distribution
Bayesian Data Analysis 3rd, Andrew Gelman et. al.
Full probability model
ri∼N(log2pCi+2(1−p)pψ+2(1−p),σri)
fi∼N(pMi+(1−p)pCi+2(1−p),σfi)
ψ=∑i(liCi)∑i(li)
- ri (log R ratio): Median-normalized log-ratio coverage of all exons within Si
- fi (B allele frequency): MAF of SNPs within segment Si
- p: Tumor purity
- Si: Genomic segment
- li: Length of $S_i
- Ci: Copy number of Si
- Mi: Copy number of minor alleles in Si, 0≤Mi≤Si
- ψ: Tumor ploidy of the sample
Sequenza results
| chromosome | start.pos | end.pos | Bf | N.BAF | sd.BAF | depth.ratio | N.ratio | sd.ratio | CNt | A | B | LPP |
|---|
chromosome | start.pos | end.pos | Bf | N.BAF | sd.BAF | depth.ratio | N.ratio | sd.ratio | CNt | A | B | LPP | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 1 | 881992 | 54694219 | 0.5 | 1636 | 0.1 | 1.1 | 2172 | 0.6 | 2 | 1 | 1 | -6.8 |
| 2 | 1 | 54700724 | 60223464 | 0.3 | 73 | 0.1 | 1.5 | 101 | 1.5 | 3 | 2 | 1 | -6.7 |
| 3 | 1 | 60381518 | 67890614 | 0 | 94 | 0 | 1.2 | 114 | 0.5 | 2 | 2 | 0 | -6.8 |
| 4 | 1 | 68151686 | 92262955 | 0.5 | 264 | 0.1 | 2.1 | 338 | 1.6 | 4 | 2 | 2 | -6.8 |
| 5 | 1 | 92445264 | 118165373 | 0.3 | 315 | 0.1 | 1.6 | 434 | 1 | 3 | 2 | 1 | -6.7 |
Sequenza results
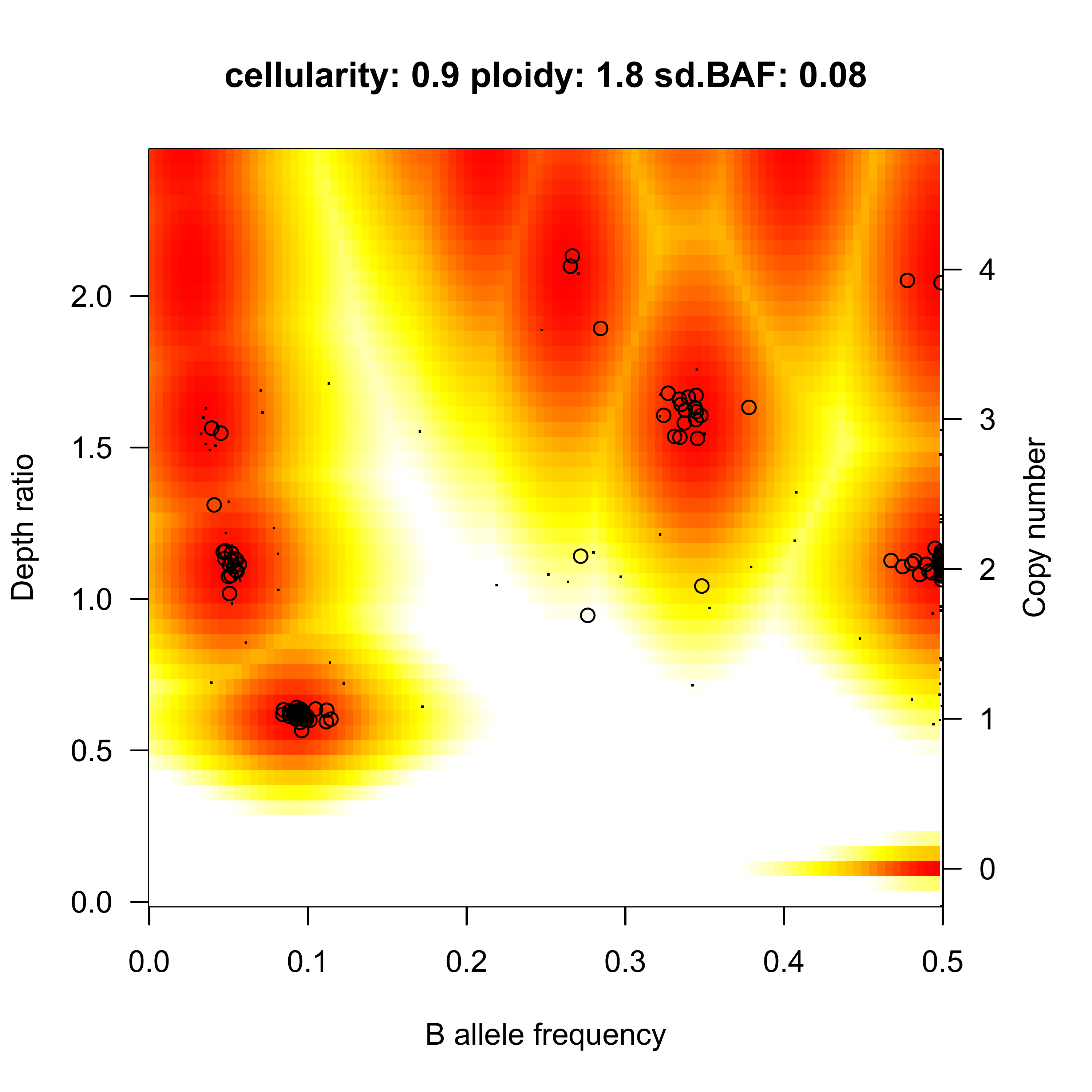
Sequenza results
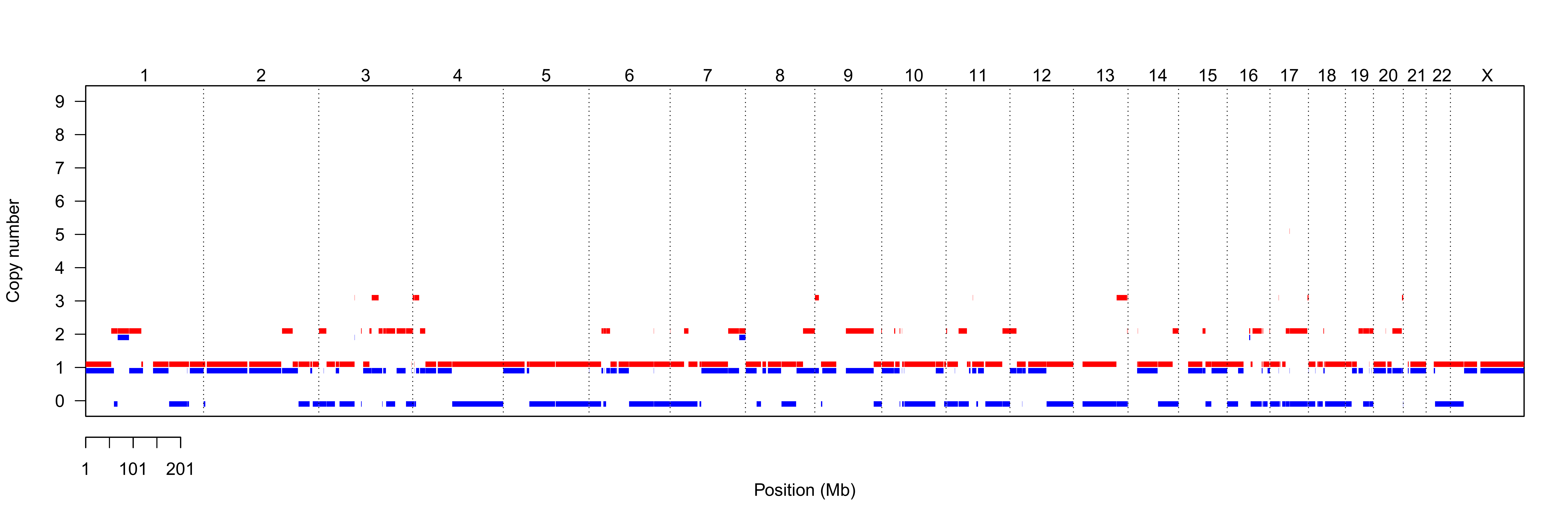
Sequenza results
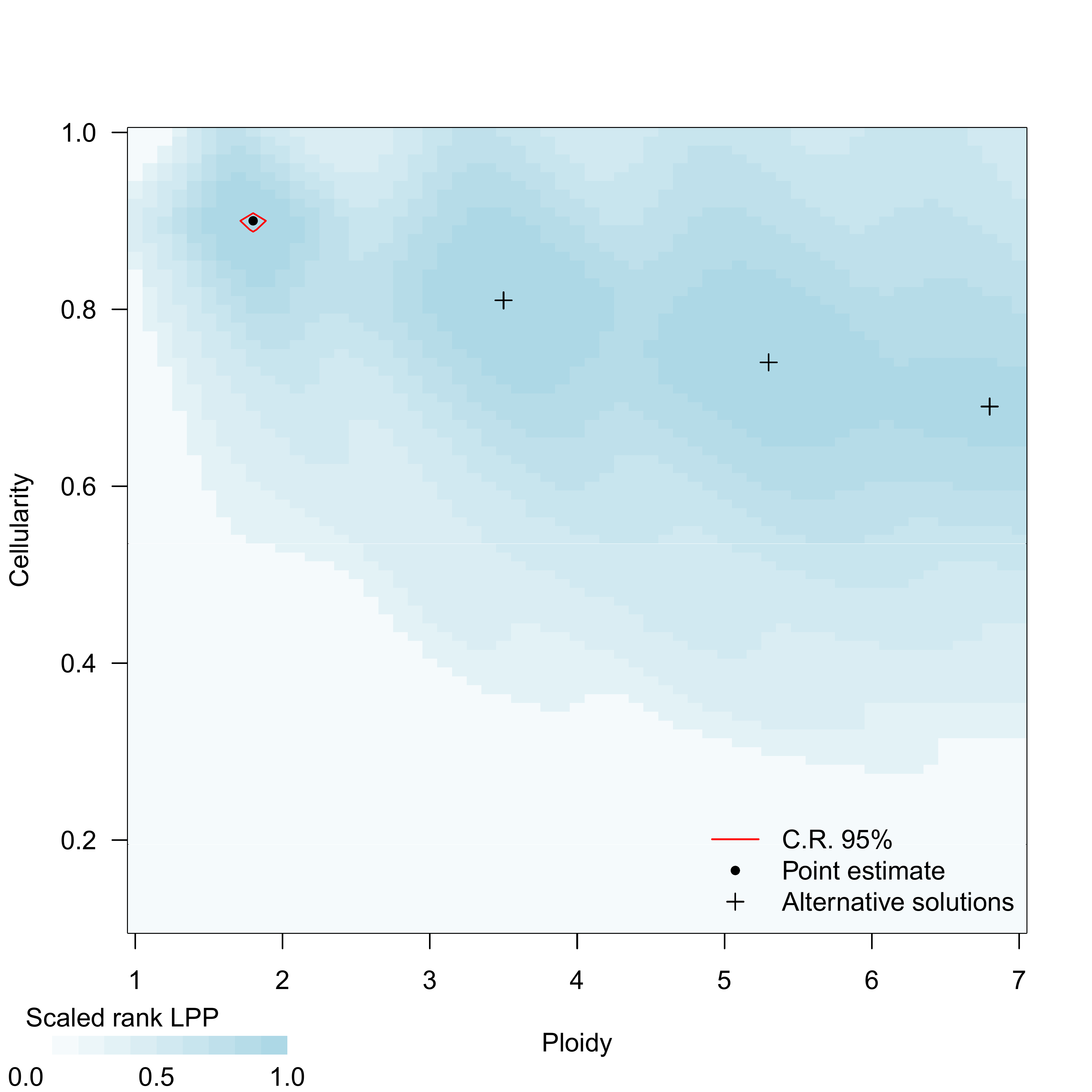
LPP: log-posterior probability
scarHRD
library(scarHRD)scar_score(here::here("static/slide/scarHRD/example/example1.txt"), reference = "grch38", chr.in.names = FALSE, seqz = FALSE)## Determining HRD-LOH, LST, TAI## HRD Telomeric AI LST HRD-sum## [1,] 27 27 33 87References
[1] V. Abkevich, K. M. Timms, B. T. Hennessy, et al. "Patterns of Genomic Loss of Heterozygosity Predict Homologous Recombination Repair Defects in Epithelial Ovarian Cancer". En. In: British Journal of Cancer 107.10 (11. 2012), pp. 1776-1782. ISSN: 1532-1827.
[2] N. J. Birkbak, Z. C. Wang, J. Kim, et al. "Telomeric Allelic Imbalance Indicates Defective DNA Repair and Sensitivity to DNA-Damaging Agents". En. In: Cancer Discov 2.4 (4. 2012), pp. 366-375. ISSN: 2159-8274, 2159-8290.
[3] T. Popova, E. Manie, G. Rieunier, et al. "Ploidy and Large-Scale Genomic Instability Consistently Identify Basal-like Breast Carcinomas with BRCA1/2 Inactivation". En. In: Cancer Res 72.21 (11. 2012), pp. 5454-5462. ISSN: 0008-5472, 1538-7445.
[4] I. Ray-Coquard, P. Pautier, S. Pignata, et al. "Olaparib plus Bevacizumab as First-Line Maintenance in Ovarian Cancer". In: New England Journal of Medicine 381.25 (12. 19, 2019), pp. 2416-2428. ISSN: 0028-4793. pmid: 31851799.
[5] F. Favero, T. Joshi, A. M. Marquard, et al. "Sequenza: Allele-Specific Copy Number and Mutation Profiles from Tumor Sequencing Data". In: Annals of Oncology 26.1 (1. 01, 2015), pp. 64-70. ISSN: 0923-7534, 1569-8041. pmid: 25319062.
[6] Z. Sztupinszki, M. Diossy, M. Krzystanek, et al. "Migrating the SNP Array-Based Homologous Recombination Deficiency Measures to next Generation Sequencing Data of Breast Cancer". In: npj Breast Cancer 4.1 (1 7. 02, 2018), pp. 1-4. ISSN: 2374-4677.
[7] E. F. Attiyeh, S. J. Diskin, M. A. Attiyeh, et al. "Genomic Copy Number Determination in Cancer Cells from Single Nucleotide Polymorphism Microarrays Based on Quantitative Genotyping Corrected for Aneuploidy". In: Genome Res. 19.2 (2. 01, 2009), pp. 276-283. ISSN: 1088-9051, 1549-5469. pmid: 19141597.
[8] J. X. Sun, Y. He, E. Sanford, et al. "A Computational Approach to Distinguish Somatic vs. Germline Origin of Genomic Alterations from Deep Sequencing of Cancer Specimens without a Matched Normal". In: PLOS Computational Biology 14.2 (2018), p. e1005965. ISSN: 1553-7358.